AFNI Tutorial #7: Group Analysis
Overview
Our goal in analyzing this dataset is to generalize the results to the population that the sample was drawn from. In other words, if we see changes in brain activity in our sample, can we say that these changes would likely be seen in the population as well?
To test this, we will run a group-level analysis (also known as a second-level analysis). In AFNI, this means that we calculate the standard error and the mean for a contrast estimate, and then test whether the average estimate is statistically significant. We will be doing this group-level analysis in two ways: Using 3dttest, which uses only the contrast estimates in testing for statistical significance; and using 3dMEMA, which accounts for both the difference between the parameter estimates, and the variability of that contrast.
Using uber_ttest.py
Just as we used a graphical user interface to preprocess the data, we can also use a GUI to set up our group-level analyses. If you type the command uber_ttest.py from the command line and press return, you will see this:
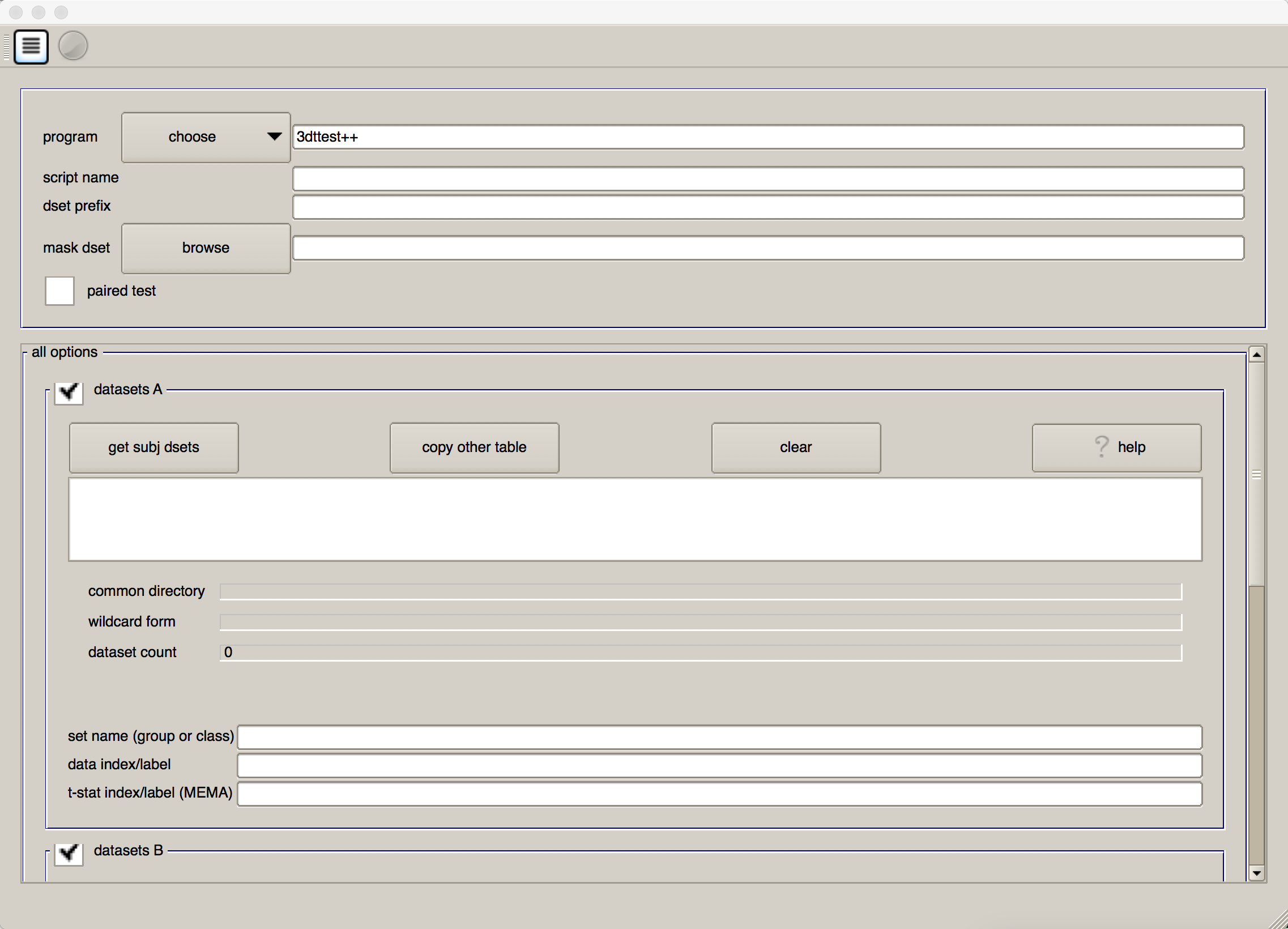
The first field, “program”, allows you to choose between 3dttest and 3dMEMA. As noted above, 3dMEMA allows you to account for the variability of the estimate as well, in order to give more weight to those subjects who have lower variability in their estimates. For now, leave it as “3dttest”.
In both the “script name” and “dset prefix” fields, enter Flanker_Inc-Con_ttest. For the “mask dset”, you can choose any one of the subject’s mask_group+tlrc images located in their results directories, as they should be similar. If you want to be more rigorous, you can calculate an intersection of the masks using AFNI’s 3dmask_tool command. The command would look something like this:
3dmask_tool -input <path/to/masks/> -prefix mask_intersection+tlrc -union
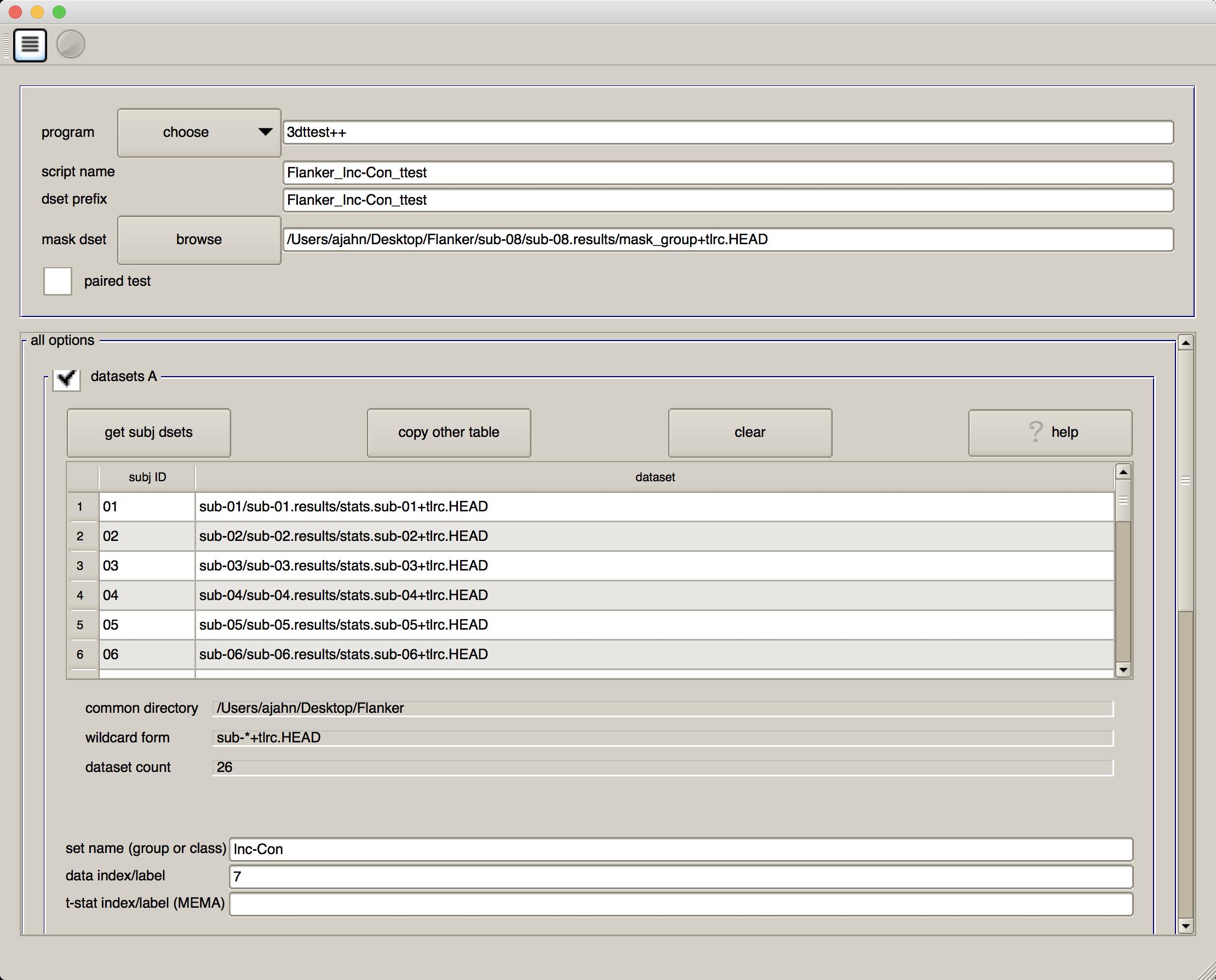
We will now select each of the subject statistical datasets, using wildcards. If you click on the button “get subj dsets” in the “datasets A” section, you will be prompted to select a representative stats dataset. Select any of the subjects’ statistical datasets, and replace the last two numbers of the subject ID with two question marks (i.e., ??). Then click on “apply pattern”. If all of the subjects were analyzed the same way and have the same directory structure, you should see 26 entries in the field below. Review them to make sure they are all there, and then click “OK”.

At the bottom of the “datasets A” section, you will see a few additional fields. In “set name (group of class), write Inc-Con, and in “data index/label” type 7.
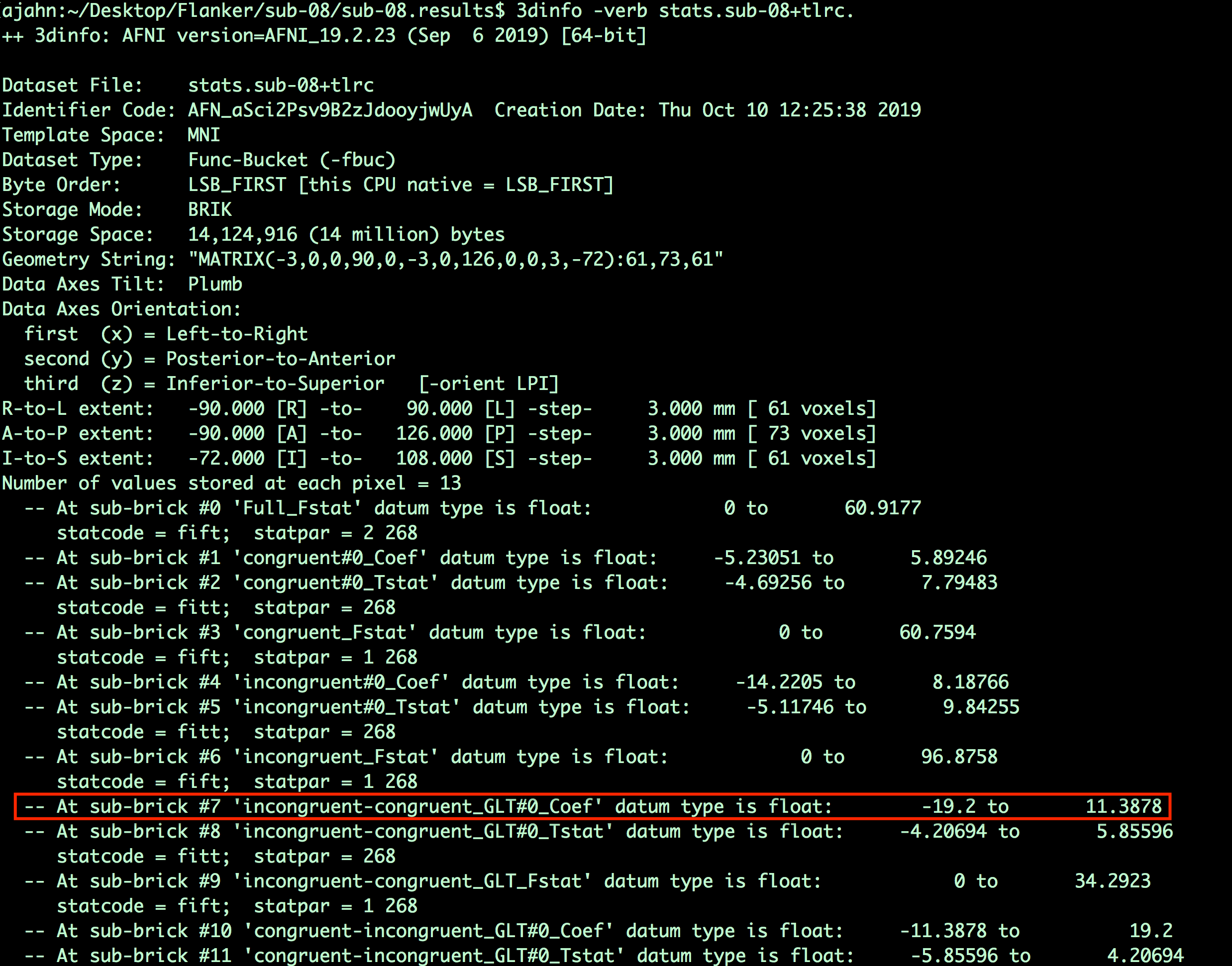
Why 7? If you open a Terminal and navigate to any of the subjects’ results directories (for example, sub-08), type
3dinfo -verb stats.sub-08+tlrc
This will show all of the sub-briks in that dataset, along with their labels. In our current example, we are looking for the contrast estimates for incongruent-congruent. The output of 3dinfo shows that this is located in sub-brik 7. Any sub-brik that contains the label “Coef” means that it is a parameter (or contrast) estimate; the label “Tstat” indicates that it is a t-statistic. (Likewise, “Fstat” means that it is an F-statistic.)
Note
What do you think the number to the right mean? For example, -19.2 to 11.3878? Why are they different between the “Coef” and the “Tstat” sub-briks? Does this difference make sense?
When you have finished, your GUI should look like the one below:
Note
If you receive an error saying that one of the fields hasn’t been filled in - but you can clearly see that it has been filled in - click on another field, and then try clicking the “Go” icon again.
Generating the Results
As with the uber_subject.py script, there are buttons at the top of the GUI for both generating the script and then running the script. First click on the icon that looks like a sheet of paper with lines on it, which will show you the command that has been generated. Review it to see how it has inserted all of your inputs into a command called 3dttest++, which will run the actual group-level analysis. Then click on the green “Go” icon to run the test. (It should take only a second.)
When it has finished, go back to your Terminal and type ls. You will see a new directory called group_results, and within that a folder called test.001.3dttest++. Navigate into that folder, which contains the script that was used to generate the results (“Flanker_Inc-Con_ttest”), and another folder called test.results, which contains the group-level output “Flanker_Inc-Con_ttest+tlrc”. Load this in the afni viewer, and overlay it on top of the MNI152 template. Threshold the images to an uncorrected p-value of 0.001 (by right-clicking on the “p=” underneath the slider bar) and clusterize the data to only show clusters with an extent of 40 voxels or more; this will create an image like the one below. Does the location of the activation make sense, given the task and the paper this experiment was based on?

Modifying the 3dttest++ Script
You can also modify the Flanker_Inc-Con_ttest script that contains the 3dttest++ command, which looks like this:
#!/bin/tcsh -xef
# created by uber_ttest.py: version 2.0 (December 28, 2017)
# creation date: Wed Feb 19 11:33:21 2020
# ---------------------- set process variables ----------------------
set mask_dset = /Users/ajahn/Desktop/Flanker/sub-01/sub-01.results/mask_group+tlrc
set dirA = /Users/ajahn/Desktop/Flanker
# specify and possibly create results directory
set results_dir = test.results
if ( ! -d $results_dir ) mkdir $results_dir
# ------------------------- process the data -------------------------
3dttest++ -prefix $results_dir/Flanker-Inc-Con \
-mask $mask_dset \
-setA Inc-Con \
01 "$dirA/sub-01/sub-01.results/stats.sub-01+tlrc[7]" \
02 "$dirA/sub-02/sub-02.results/stats.sub-02+tlrc[7]" \
03 "$dirA/sub-03/sub-03.results/stats.sub-03+tlrc[7]" \
04 "$dirA/sub-04/sub-04.results/stats.sub-04+tlrc[7]" \
05 "$dirA/sub-05/sub-05.results/stats.sub-05+tlrc[7]" \
06 "$dirA/sub-06/sub-06.results/stats.sub-06+tlrc[7]" \
07 "$dirA/sub-07/sub-07.results/stats.sub-07+tlrc[7]" \
08 "$dirA/sub-08/sub-08.results/stats.sub-08+tlrc[7]" \
09 "$dirA/sub-09/sub-09.results/stats.sub-09+tlrc[7]" \
10 "$dirA/sub-10/sub-10.results/stats.sub-10+tlrc[7]" \
11 "$dirA/sub-11/sub-11.results/stats.sub-11+tlrc[7]" \
12 "$dirA/sub-12/sub-12.results/stats.sub-12+tlrc[7]" \
13 "$dirA/sub-13/sub-13.results/stats.sub-13+tlrc[7]" \
14 "$dirA/sub-14/sub-14.results/stats.sub-14+tlrc[7]" \
15 "$dirA/sub-15/sub-15.results/stats.sub-15+tlrc[7]" \
16 "$dirA/sub-16/sub-16.results/stats.sub-16+tlrc[7]" \
17 "$dirA/sub-17/sub-17.results/stats.sub-17+tlrc[7]" \
18 "$dirA/sub-18/sub-18.results/stats.sub-18+tlrc[7]" \
19 "$dirA/sub-19/sub-19.results/stats.sub-19+tlrc[7]" \
20 "$dirA/sub-20/sub-20.results/stats.sub-20+tlrc[7]" \
21 "$dirA/sub-21/sub-21.results/stats.sub-21+tlrc[7]" \
22 "$dirA/sub-22/sub-22.results/stats.sub-22+tlrc[7]" \
23 "$dirA/sub-23/sub-23.results/stats.sub-23+tlrc[7]" \
24 "$dirA/sub-24/sub-24.results/stats.sub-24+tlrc[7]" \
25 "$dirA/sub-25/sub-25.results/stats.sub-25+tlrc[7]" \
26 "$dirA/sub-26/sub-26.results/stats.sub-26+tlrc[7]"
Using this code as a template, you would change the directory for the output and the mask to a location on your machine. You would then replace each of the input files with the ones that you are interested in comparing, while keeping an index number next to each (e.g., 1 through 26 in this example).
If you want to compare two different groups, you would use the -setB option:
#!/bin/tcsh -xef
# created by uber_ttest.py: version 2.0 (December 28, 2017)
# creation date: Wed Feb 19 13:23:11 2020
# ---------------------- set process variables ----------------------
set mask_dset = /Users/ajahn/Desktop/Flanker/sub-01/sub-01.results/mask_group+tlrc
set dirA = /Users/ajahn/Desktop/Flanker
# specify and possibly create results directory
set results_dir = test.results
if ( ! -d $results_dir ) mkdir $results_dir
# ------------------------- process the data -------------------------
3dttest++ -prefix $results_dir/Flanker-Inc-Con -AminusB \
-mask $mask_dset \
-setA Inc-Con_Group1 \
01 "$dirA/sub-01/sub-01.results/stats.sub-01+tlrc[7]" \
02 "$dirA/sub-02/sub-02.results/stats.sub-02+tlrc[7]" \
03 "$dirA/sub-03/sub-03.results/stats.sub-03+tlrc[7]" \
04 "$dirA/sub-04/sub-04.results/stats.sub-04+tlrc[7]" \
05 "$dirA/sub-05/sub-05.results/stats.sub-05+tlrc[7]" \
06 "$dirA/sub-06/sub-06.results/stats.sub-06+tlrc[7]" \
07 "$dirA/sub-07/sub-07.results/stats.sub-07+tlrc[7]" \
08 "$dirA/sub-08/sub-08.results/stats.sub-08+tlrc[7]" \
09 "$dirA/sub-09/sub-09.results/stats.sub-09+tlrc[7]" \
10 "$dirA/sub-10/sub-10.results/stats.sub-10+tlrc[7]" \
11 "$dirA/sub-11/sub-11.results/stats.sub-11+tlrc[7]" \
12 "$dirA/sub-12/sub-12.results/stats.sub-12+tlrc[7]" \
13 "$dirA/sub-13/sub-13.results/stats.sub-13+tlrc[7]" \
-setB Inc-Con_Group2 \
14 "$dirA/sub-14/sub-14.results/stats.sub-14+tlrc[7]" \
15 "$dirA/sub-15/sub-15.results/stats.sub-15+tlrc[7]" \
16 "$dirA/sub-16/sub-16.results/stats.sub-16+tlrc[7]" \
17 "$dirA/sub-17/sub-17.results/stats.sub-17+tlrc[7]" \
18 "$dirA/sub-18/sub-18.results/stats.sub-18+tlrc[7]" \
19 "$dirA/sub-19/sub-19.results/stats.sub-19+tlrc[7]" \
20 "$dirA/sub-20/sub-20.results/stats.sub-20+tlrc[7]" \
21 "$dirA/sub-21/sub-21.results/stats.sub-21+tlrc[7]" \
22 "$dirA/sub-22/sub-22.results/stats.sub-22+tlrc[7]" \
23 "$dirA/sub-23/sub-23.results/stats.sub-23+tlrc[7]" \
24 "$dirA/sub-24/sub-24.results/stats.sub-24+tlrc[7]" \
25 "$dirA/sub-25/sub-25.results/stats.sub-25+tlrc[7]" \
26 "$dirA/sub-26/sub-26.results/stats.sub-26+tlrc[7]"
Non-Parametric Tests with 3dClustSim
Note
For details on cluster thresholding and the terms that are used in this section, see Appendix A of the fMRI Course, focusing on the section “AFNI’s 3dFWHMx and 3dClustSim”.
Newer versions of AFNI (from about 2019 onwards) have a new option, -3dClustSim. This carries out a non-parametric test which doesn’t make any assumptions about the shape of the null distribution; instead, a null distribution is created from the data itself, and statistical significance is determined by comparing the data to its own null distribution.
This would be done by inserting the string -3dClustSim after the 3dttest++ command; e.g.:
3dttest++ -ClustSim -prefix $results_dir/Flanker-Inc-Con \
-mask $mask_dset \
-setA Inc-Con
And keeping the rest of the input files the same.
The output of this command will be a z-statistic image which you can threshold as you would any other statistical image. The command will also output .1D files that contain lists of how large a cluster has to be in order to be determined significant. For example, the file TTnew.CSimA.NN3_bisided.1D will contain rows of each cluster-forming threshold and columns for each corresponding alpha threshold. For example, you may see output like this:
# CLUSTER SIZE THRESHOLD(pthr,alpha) in Voxels
# -NN 3 | alpha = Prob(Cluster >= given size)
# pthr | .10000 .09000 .08000 .07000 .06000 .05000 .04000 .03000 .02000 .01000
# ------ | ------ ------ ------ ------ ------ ------ ------ ------ ------ ------
0.100000 7659 8174 8664 9262 9840 10571 11286 12271 13609 15859
0.090000 6325 6804 7356 8015 8596 9246 10045 10982 12266 14584
0.080000 4753 5301 5845 6425 7182 7830 8580 9649 10974 13194
0.070000 3283 3696 4184 4778 5439 6243 7172 8153 9453 11699
0.060000 2000 2291 2666 3136 3676 4409 5324 6295 7808 10056
0.050000 1160 1336 1542 1797 2187 2629 3289 4323 5621 8082
0.040000 659 740 843 1005 1194 1469 1801 2281 3385 5796
0.030000 351 383 427 496 578 696 887 1148 1533 2837
0.020000 170 188 207 230 262 305 373 476 664 1097
0.015000 116 125 137 151 167 189 227 290 400 616
0.010000 72 77 82 90 99 113 129 158 195 283
0.007000 50 53 57 61 67 74 84 100 123 177
0.005000 36 38 41 44 48 53 60 69 83 117
0.003000 24 26 27 29 31 34 37 44 53 69
0.002000 18 19 20 22 23 25 28 31 37 48
0.001500 15 16 17 18 19 20 22 25 30 39
0.001000 12 13 13 14 15 16 17 19 22 29
Going down the list to the row next to a cluster-forming threshold of p=0.001, we see that we would need a cluster size of 16 or greater in order to determine that the cluster is significant at the p=0.05 threshold.
Using 3dMEMA
Close the AFNI viewer, and then use the Terminal to navigate back to the directory containing your subjects. Go back to the uber_ttest.py GUI (or open a new one), and make the following changes:
Change the “program” from 3dttest++ to 3dMEMA.
Change the “script name” and “dset prefix” to
Flanker_Inc-Con_MEMA.Click on “get subj dsets”, and select a subject’s statistical dataset that has the “REML” string (e.g.,
stats.sub-08_REML+tlrc). Use the wildcards as above to select all of the subjects’ REML datasets.In the field “t-stat index/label (MEMA)”, type
8. The sub-briks of the REML dataset, which should be in an order identical to the non-REML statistical dataset, indicate that sub-brik #8 is the t-statistic associated with the contrast estimate of “incongruent-congruent.”
As before, click on the script generator icon, and then click on the green “Go” button. This model estimation will take longer, and you will see a progress report for each slice that has been analyzed; in total, it should take only a couple of minutes.
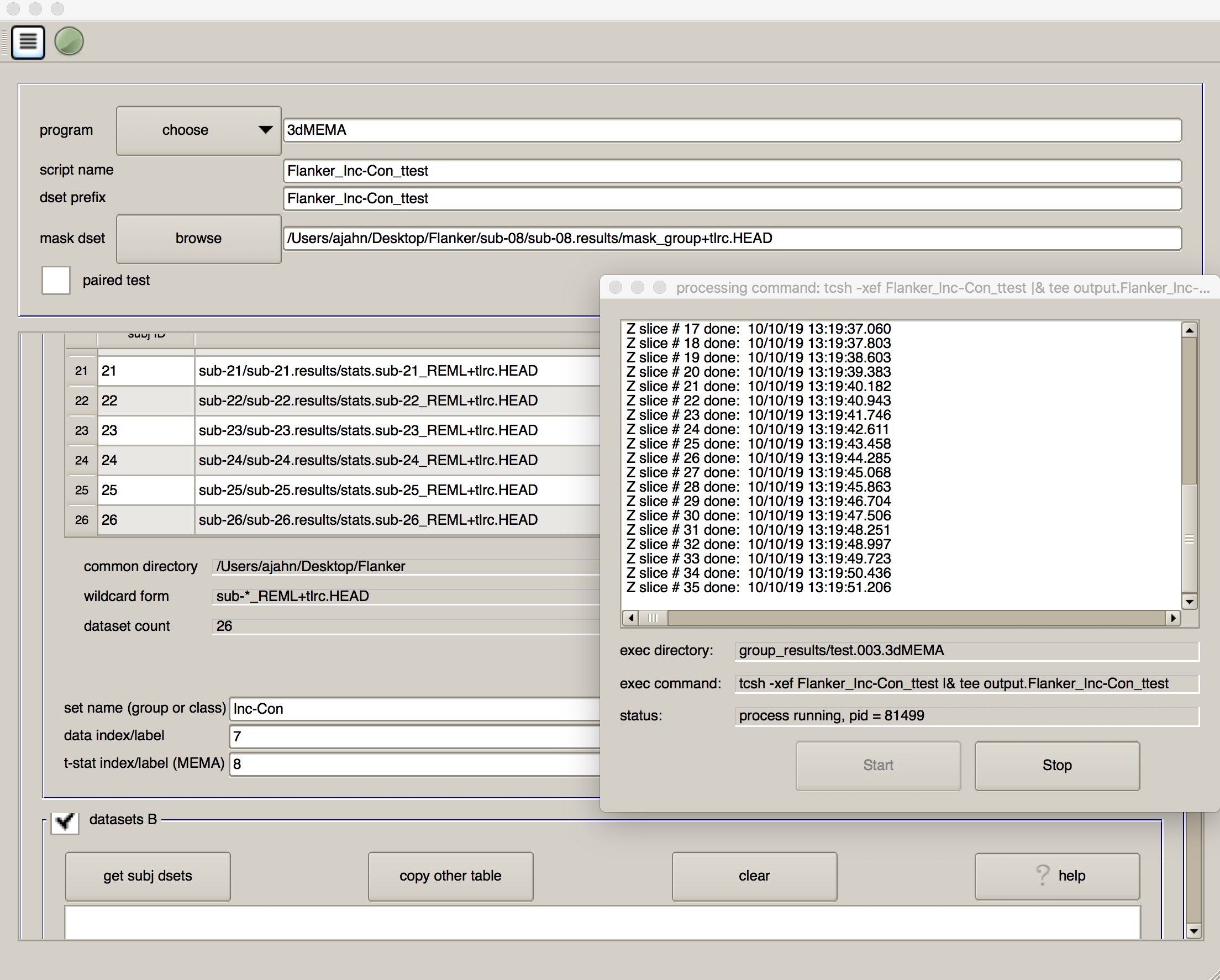
When it has finished, you will see a new directory in the group_results folder called test.002.3dMEMA, with a sub-directory called test.results. Navigate to that folder, and overlay the results as before. Are the effects in the same location? Do these effects look stronger or weaker? Why?
Video
Click here for a walkthrough of how to do group-level analysis in AFNI.